As noted in the previous chapters, Python's for loop works with collections. When working with materialized collections such as tuples, lists, maps, and sets, the for loop involves the explicit management of states. While this strays from purely functional programming, it reflects a necessary optimization for Python. If we assume that state management is localized to an iterator object that's created as a part of the for statement evaluation, we can leverage this feature without straying too far from pure, functional programming. If, for example, we use the for loop variable outside the indented body of the loop, we've strayed from purely functional programming by leveraging this state control variable.
We'll return to this in Chapter 6, Recursions and Reductions. It's an important topic, and we'll just scratch the surface here with a quick example of working with generators.
One common application of for loop iterable processing is the unwrap(process(wrap(iterable))) design pattern. A wrap() function will first transform each item of an iterable into a two-tuple with a derived sort key and the original item. We can then process these two-tuple items as a single, wrapped value. Finally, we'll use an unwrap() function to discard the value used to wrap, which recovers the original item.
This happens so often in a functional context that two functions are used heavily for this; they are the following:
fst = lambda x: x[0] snd = lambda x: x[1]
These two functions pick the first and second values from a two-tuple, and both are handy for the process() and unwrap() functions.
Another common pattern is wrap3(wrap2(wrap1())). In this case, we're starting with simple tuples and then wrapping them with additional results to build up larger and more complex tuples. A common variation on this theme builds new, more complex namedtuple instances from source objects. We can summarize both of these as the Accretion design pattern—an item that accretes derived values.
As an example, consider using the accretion pattern to work with a simple sequence of latitude and longitude values. The first step will convert the simple point represented as a (lat, lon) pair on a path into pairs of legs (begin, end). Each pair in the result will be represented as ((lat, lon), (lat, lon)). The value of fst(item) is the starting position; the value of snd(item) is the ending position for each value of each item in the collection.
In the next sections, we'll show you how to create a generator function that will iterate over the content of a file. This iterable will contain the raw input data that we will process.
Once we have the raw data, later sections will show how to decorate each leg with the haversine distance along the leg. The final result of a wrap(wrap(iterable())) design will be a sequence of three tuples—((lat, lon), (lat, lon), distance). We can then analyze the results for the longest and shortest distance, bounding rectangle, and other summaries.























































 , we would also want to create a paired sequence,
, we would also want to create a paired sequence,  . The first and second items form a pair. The second and third items form the next pair. When doing time-series analysis, we may be combining more widely separated values. In this example, the pairs are immediately adjacent values.
. The first and second items form a pair. The second and third items form the next pair. When doing time-series analysis, we may be combining more widely separated values. In this example, the pairs are immediately adjacent values.



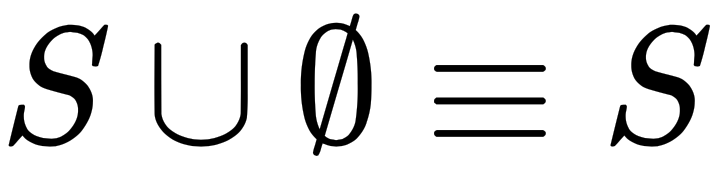 . If we union a set, S, with an empty set, we get the original set, S. The empty set can be called the union identify element. This is parallel to the way zero is the additive identity element:
. If we union a set, S, with an empty set, we get the original set, S. The empty set can be called the union identify element. This is parallel to the way zero is the additive identity element:
 , then all(()) must be True.
, then all(()) must be True. .
. .
.













